9.8. Archive view
9.8.1. Background
This view provides an “archive” (read-only) view of an electronic health record (EHR). It has two potential purposes:
to provide a useful, configurable way in which researchers can explore the anonymised EHR of a single patient;
to act as an archive view onto an identifiable EHR, if so configured.
It is entirely configurable by the local system administrators, though CRATE comes with some specimen archive views.
9.8.2. How to customize the archive view
9.8.2.1. Write your archive
Within a directory tree of your choice, write Mako templates. Some specimen miniature web sites are provided to show you how, in:
crate_anon/crateweb/specimen_archives/basic/
crate_anon/crateweb/specimen_archives/tree/
The “basic” one demonstrates basic layout, SQL queries, and downloading of binary attachments. The “tree” one uses a collapsible tree-style menu on the left and a range of specimen views onto the EHR on the right.
This system gives you full use of HTML/Javascript and Python simultaneously. (Python code will run within the same interpreter and virtual environment used by CRATE.)
There are assistance functions that you can import, such as those in
crate_anon.crateweb.research.archive_func.
9.8.2.2. The archive’s Python context
Mako templates have a context, which is a collection of Python objects “visible” to template code (see Mako Runtime Environment). In the CRATE archive, this context is built up as follows:
The config file’s ARCHIVE_CONTEXT dictionary is shallow-copied with
copy.copy(). You can use ARCHIVE_CONTEXT to pass a set of custom variables to your templates.That copy is updated with a specific set of keys, described next, which become visible as Python objects. (Doing it in this order means that ARCHIVE_CONTEXT can’t override the special CRATE keys.)
Mako will later add a few special objects of its own (see Mako Runtime Environment).
The special objects provided by CRATE are:
CRATE_HOME_URL:URL to your site’s CRATE home page. Use this to escape from the archive view!
execute:Function to run an SQL query (via the research database connection), or just execute raw SQL, and return a database cursor. Call it as
cursor = execute(sql) cursor = execute(sql, args)
Use question marks (
?) in the SQL as argument placeholders.get_attachment_url:Function to generate a URL to a binary attachment. This function adds the referring patient ID (for audit purposes) and calls
crate_anon.crateweb.research.views.archive_attachment_url()(see that for details). Call it like this:get_attachment_url(filename, ...)
get_static_url:Function to generate a URL to a binary attachment, which is
crate_anon.crateweb.research.views.archive_static_url()(see that for details). Call it like this:get_static_url(filename, ...)
get_patient_template_url:Function to generate a URL to a template in another part of the archive, for the same patient. Call it as
get_patient_template_url(template_name, **kwargs)
You can pass any keyword parameters except:
patient_idtemplatemtime
get_template_url:Function to generate a URL to a template in another part of the archive, without (necessarily) passing a patient ID. Call it as
get_template_url(template_name, **kwargs)
You can pass any keyword parameters except:
templatemtime
patient_id:The ID of this patient. (A string, but that will still work an an SQL parameter for integer fields. You can of course process it further if you wish; this is illustrated in the “tree” example.)
query_params:The HTTP GET query parameters, as a Django
django.http.request.QueryDict.request:The Django HTTP request, a
django.http.request.HttpRequestobject.
These objects are directly accessible, e.g. as ${patient_id}, in Mako
statements. In the more obvious Python blocks (e.g. within Mako’s <% ... %>
blocks), they are also accessible, as (in this example) any of patient_id,
context["patient_id"], or context[ArchiveContextKeys.patient_id].
One use for ARCHIVE_CONTEXT is to develop a set of templates that operate either with an original identified clinical records database or with a de-identified version with slightly different structure (but similar enough to want to avoid code redundancy). You could set a flag in ARCHIVE_CONTEXT to tell your templates which one is currently in use.
9.8.2.3. Point CRATE at your archive
See the relevant section of the web config file.
9.8.3. Examples
Here’s part of the demonstration tree-style archive, with entirely fictional data (and de-identified to boot).
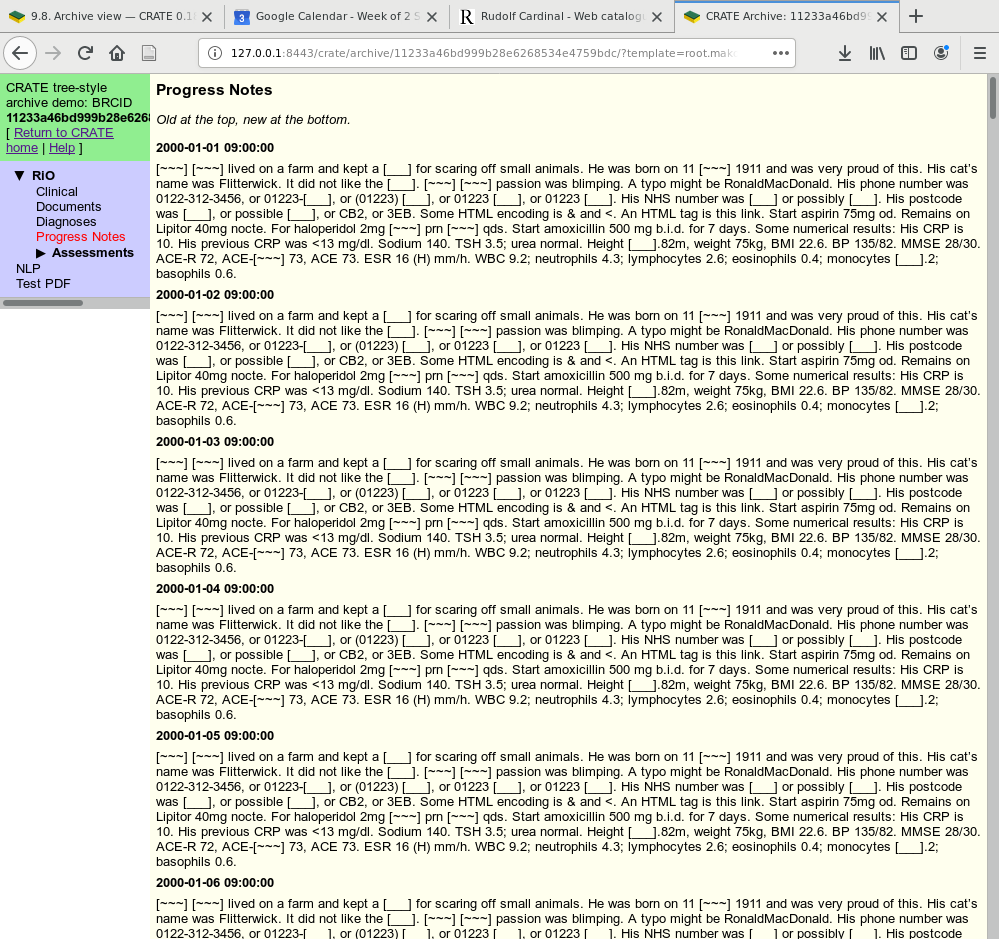
“Progress Notes” display. The template has fetched data for the current patient via an SQL query and reformatted it to look like a conventional EHR “progress notes” journal (though in this case without author information).
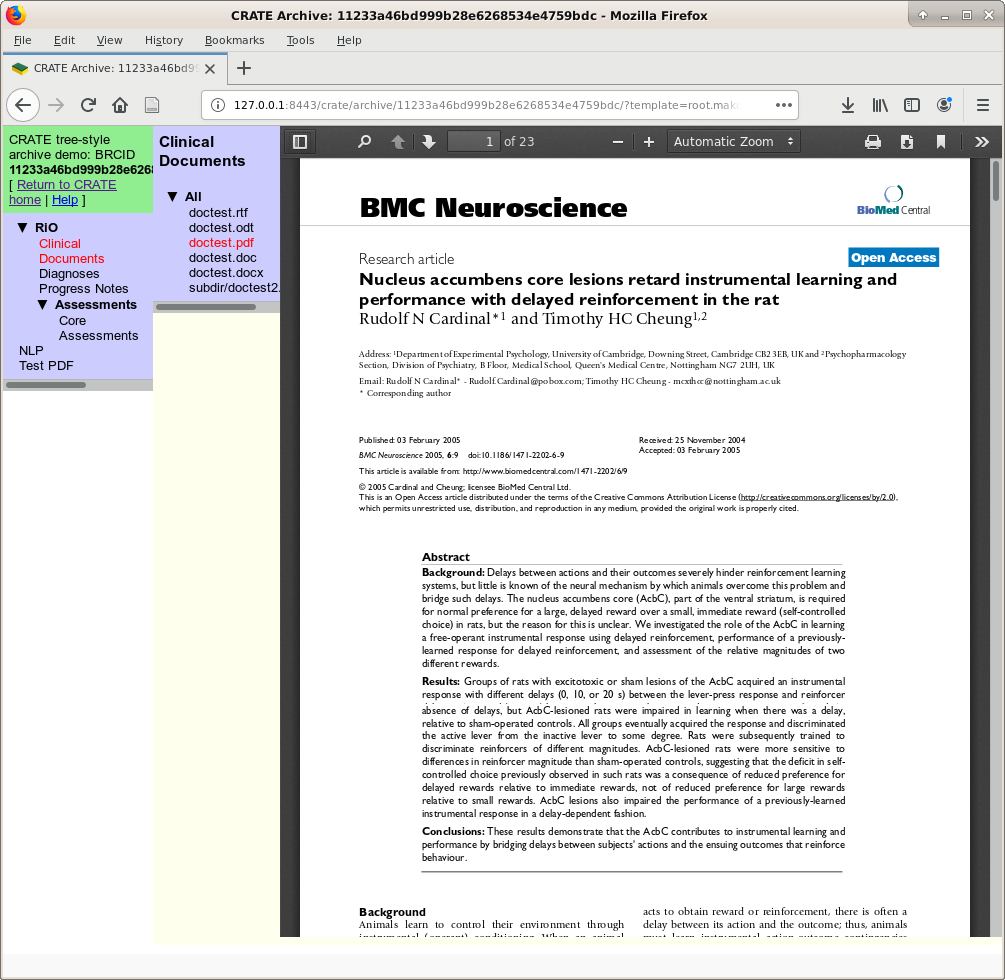
“Clinical Documents” display, showing PDFs inline.
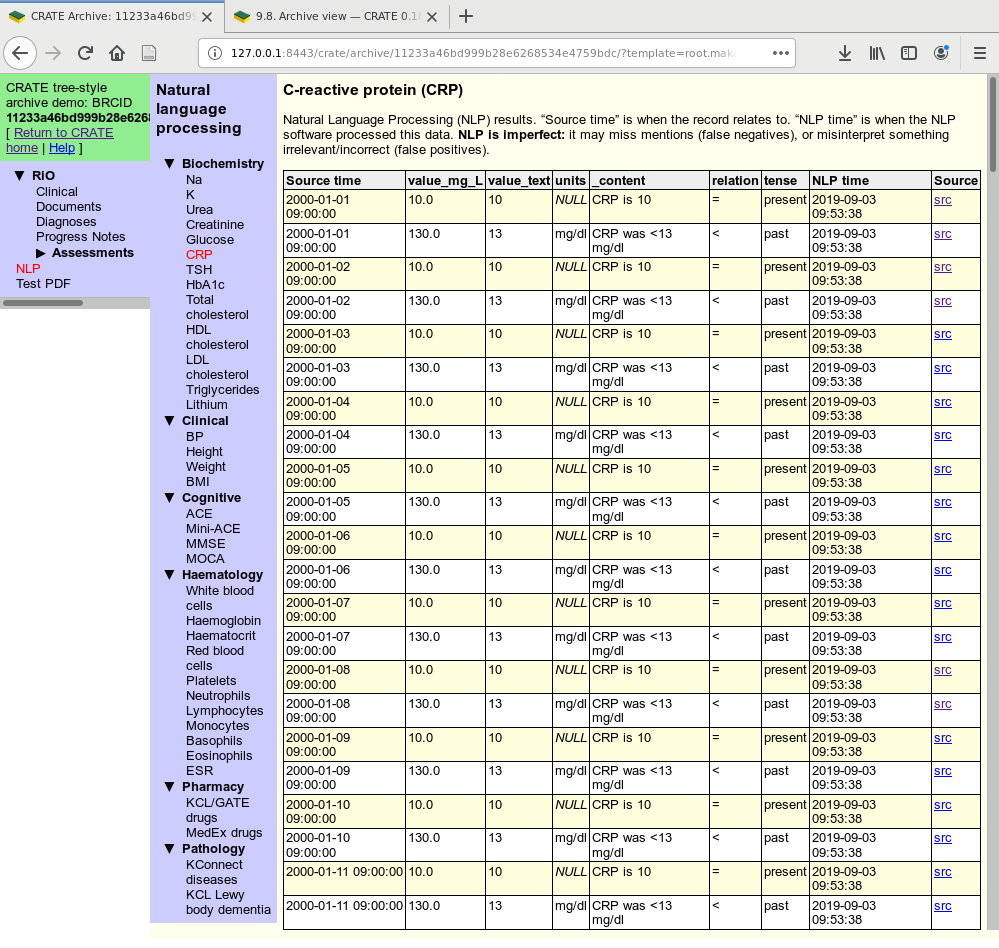
View on a NLP table, created by CRATE through analysis of free text. A generic “query results” template is used.
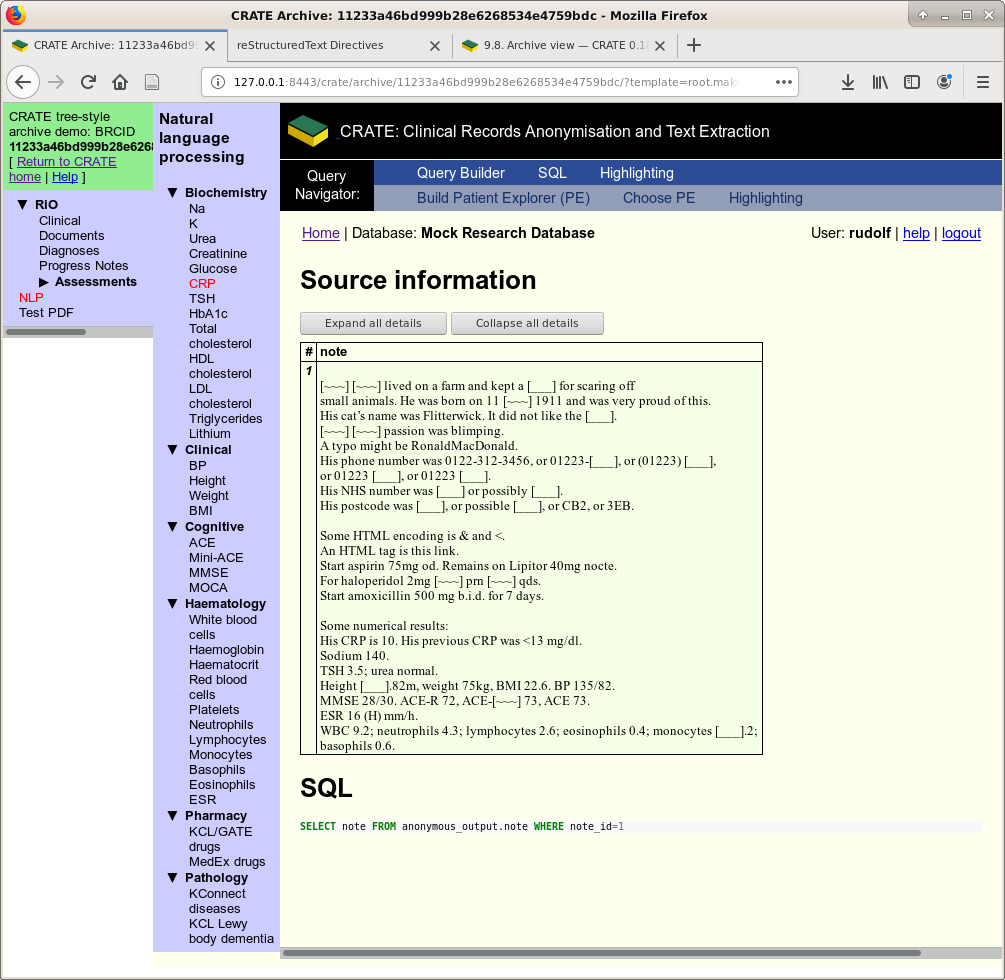
The NLP results hyperlink through to their source data, if available. Here’s the note that generated one of the CRP values.
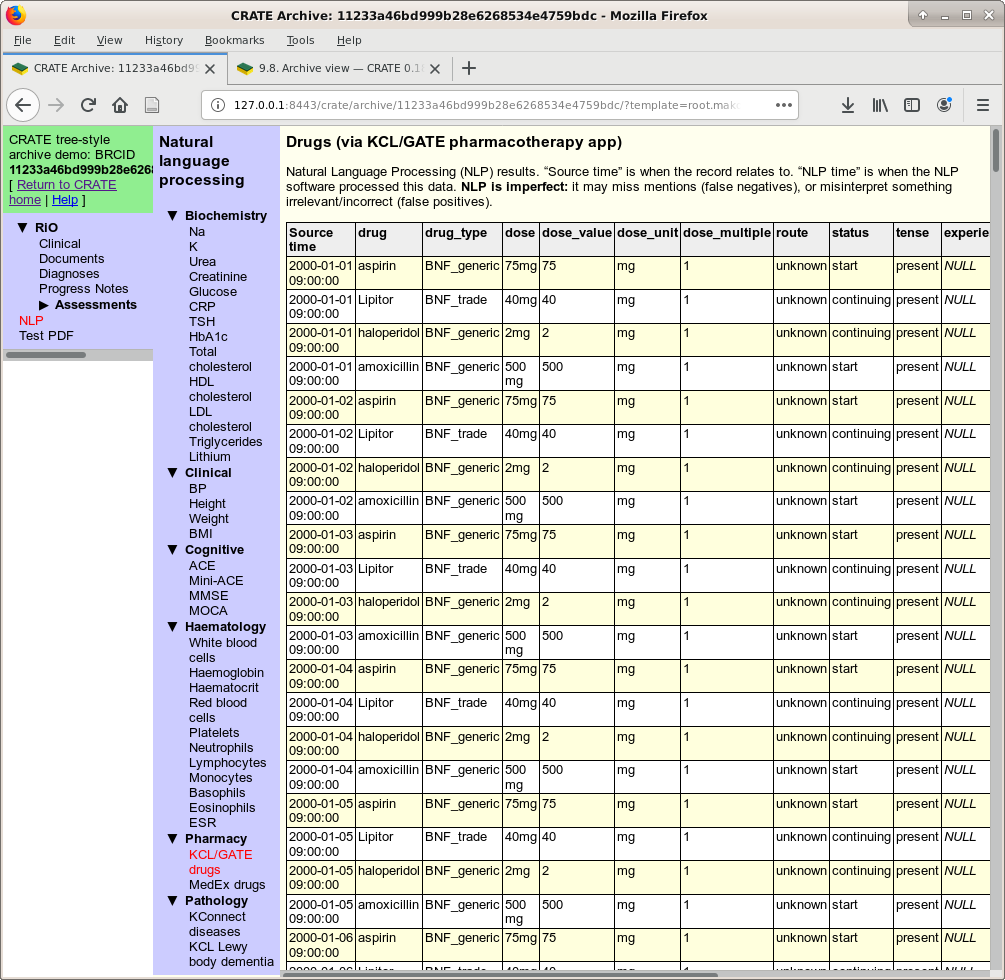
Another NLP view, this time of drugs found via the KCL GATE pharmacotherapy app.
9.8.4. Design notes
2019-08-31:
HTML templates, written locally, stored on disk in a user-defined directory.
Any template engine would be reasonable, but the two obvious candidates are
Django, because we use that for the CRATE web front end (but the template language is somewhat restricted);
Mako, because the templates can include arbitrary Python, and because Django/Mako interoperability is possible (including via Django-Mako-Plus but also directly).
Other template engines, but nothing is particularly compelling over those two.
Let’s use Mako.
A structure that is configurable by the local administrator (stored in a config file, a database, or on disk), mapping the templates.
The best is probably to specify a single template as the root template in the config file.
A URL system to produce requests to other parts of the archive, with arbitrary parameters via HTTP GET URL parameters.
Pre-population of the template dictionary with useful objects (but not those that take much time to create). See
crate_anon.crateweb.research.views.archive_template().
Not done:
consider Windows authentication to Django
optional launch page for archive (e.g. allowing JSON POST for patient ID)