1. Overview
A description is provided in Cardinal (2017) “Clinical records anonymisation and text extraction (CRATE): an open-source software system”, BMC Medical Informatics and Decision Making 17: 50; PubMed ID 28441940; http://doi.org/10.1186/s12911-017-0437-1.
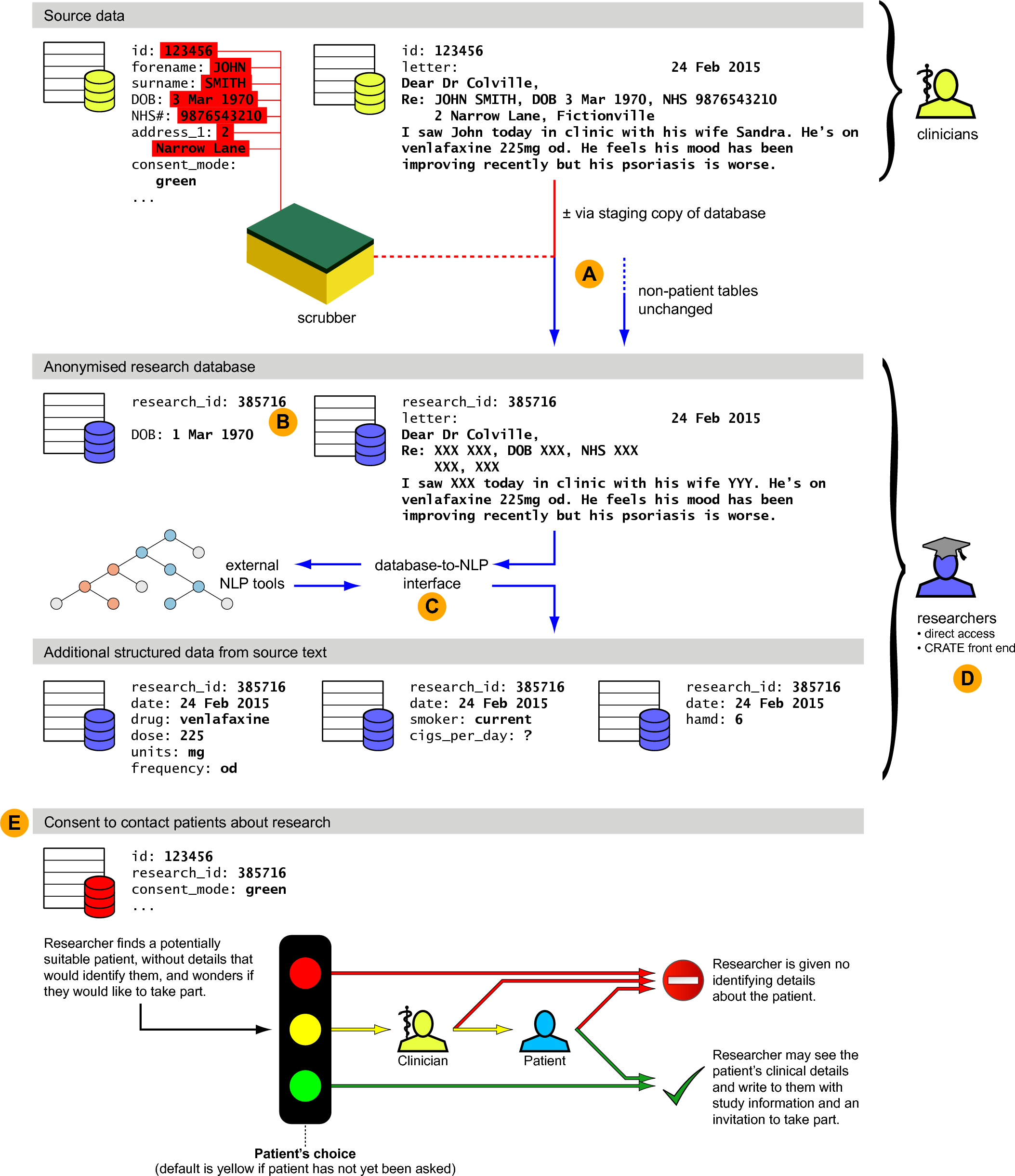
Overview of the roles that CRATE can play in the creation of a research database. The figure shows a schematic of a full EMR containing sensitive and identifiable information, its processing into a pseudonymised research database, and methods through which researchers may use the research database to contact patients about research, while preserving anonymity for those who have not consented to be contacted. Key functions of CRATE are shown, as follows. (A) Anonymisation of source data in a relational database framework, using identifiers in the source data to ‘scrub’ free text. In this example the date of birth has also been partially obscured. (B) Generation of crypographically secure research IDs using hashed message authentication codes and one-time pads. An integer transient research ID is illustrated; full research IDs use longer hexadecimal digests. (C) Provision of a managed relational database interface to natural language processing tools such as GATE. (D) Provision of an optional web front end to a research database. (E) Management of a consent-to-contact process. The anonymisation, NLP, front end, and consent-to-contact components are modular and usable separately.
Source code is at https://github.com/ucam-department-of-psychiatry/crate.
CRATE is https://web.www.healthdatagateway.org/tool/5247022928209417 at the HDR UK Innovation Gateway.